Signatures
Cell Types
Publications
Tissues
Diseases
Species
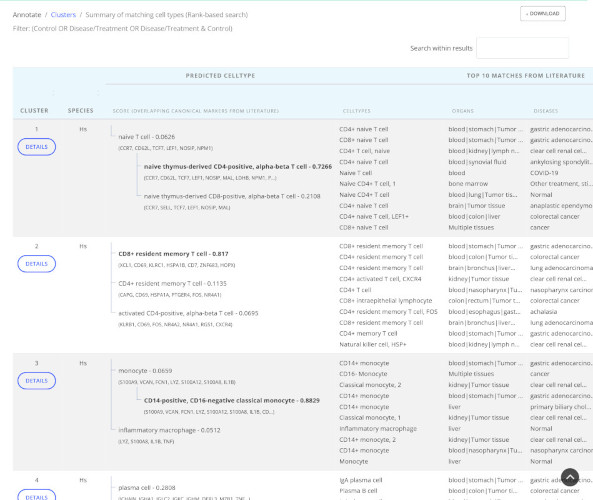
Cluster annotation
Predict cell types for cluster-specific gene lists.
Single-cell annotation
Annotate individual cells in scRNA-seq datasets.
Spatial deconvolution
Assign cell types in spatial RNA-seq datasets.
Bulk cell type enrichment
Find cell type abundance in bulk RNA-seq data.
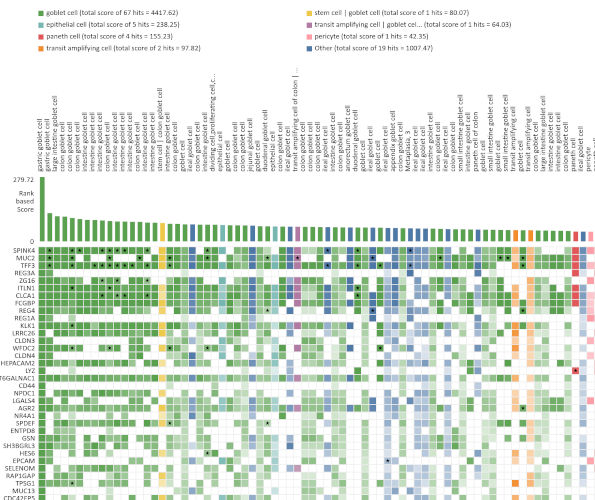
Consensus signatures
View consensus signatures for cell types in disease.
Biomarker discovery
Get top marker genes for cell types in tissues.
High-quality in-built reference
Large number of cell types, tissues, diseases and species available in a single, curated resource.
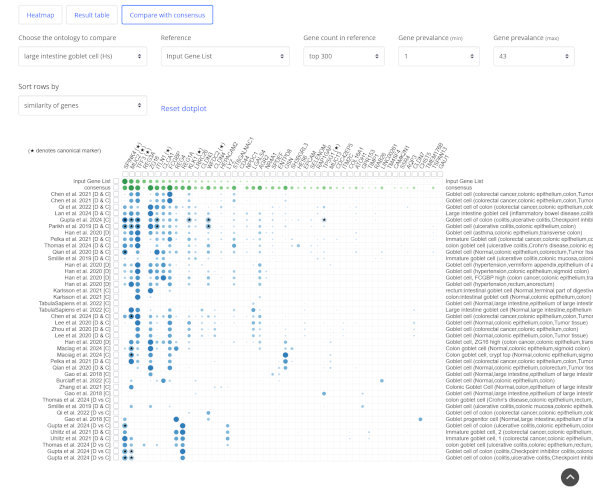
Marker based annotation
Fast rank-based matching method compares upregulated genes with every cell type signature from all references independently.
Granular cell type predictions
Get broad and granular predicted cell types with publications and previously used canonical markers for confirmation.
AI-ready annotations
Standardized ontologies for cell types, tissues and diseases for AI-ready data labeling.
Search across organisms
Annotate cell types in non-model organisms using reference data from model organisms.
Regular updates
Our database is updated with the latest publications every 4 months.