Cell type annotation of clusters
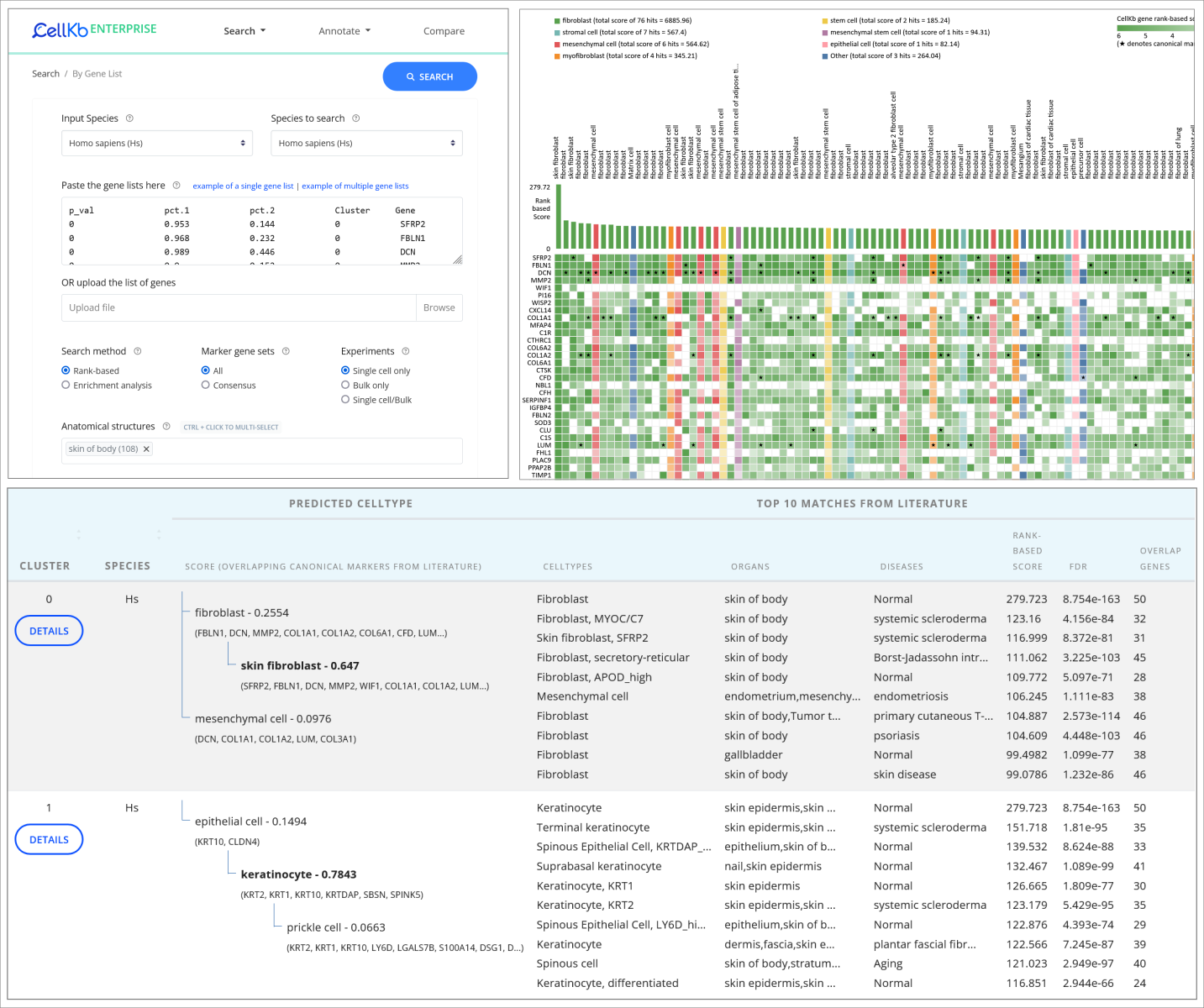
Single-cell annotation
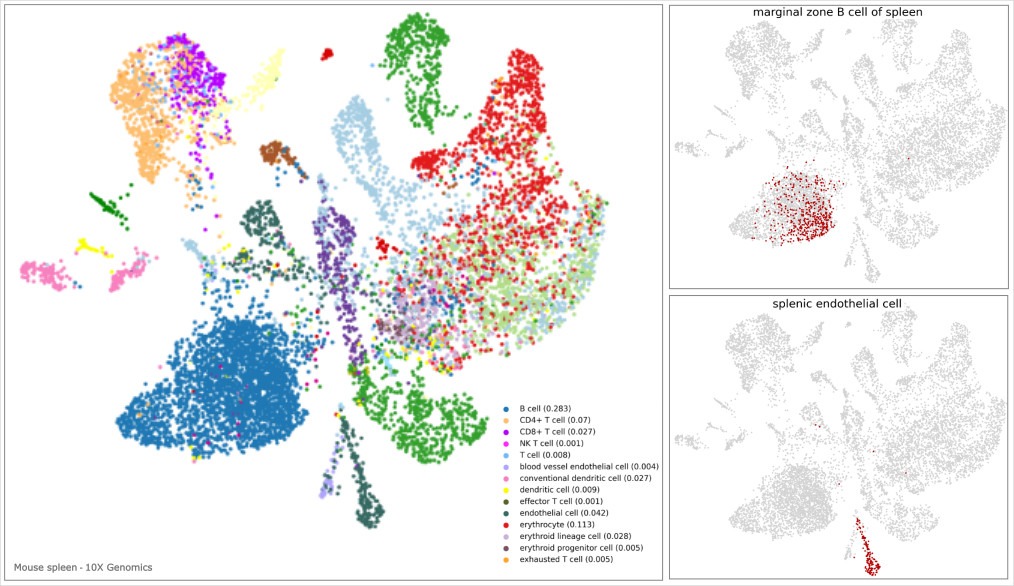
Spot deconvolution
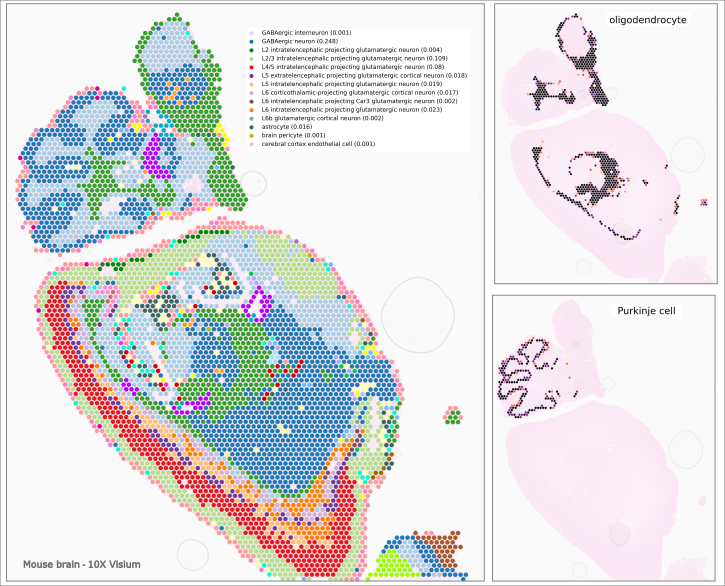
Contact us for a trial to the scRNA-seq and spatial RNA-seq data annotation service in CellKb.
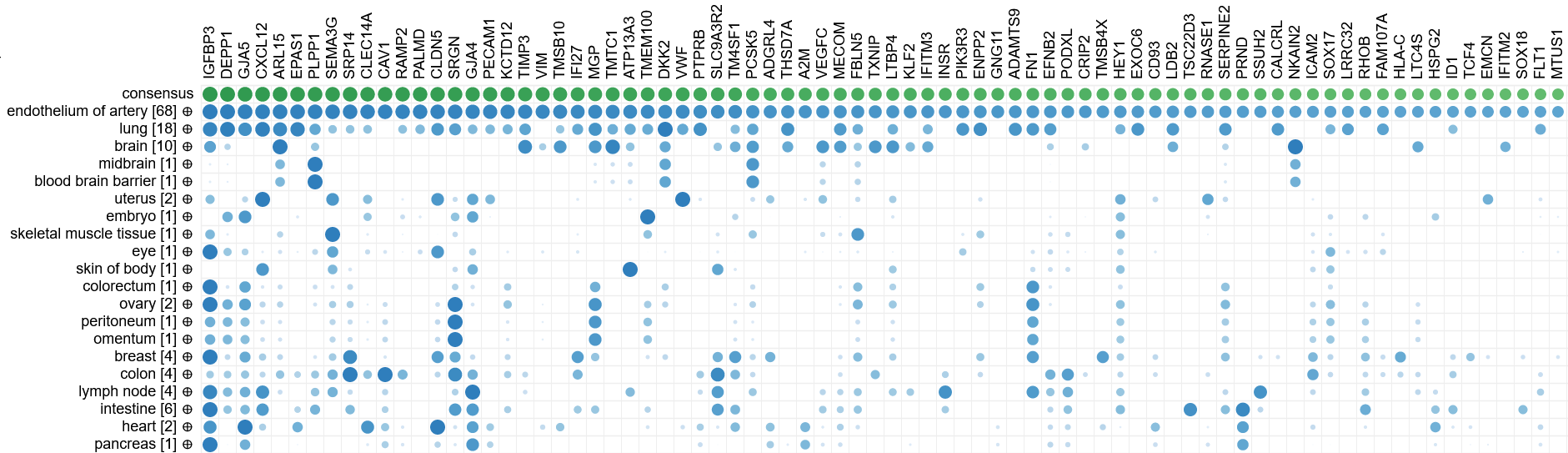
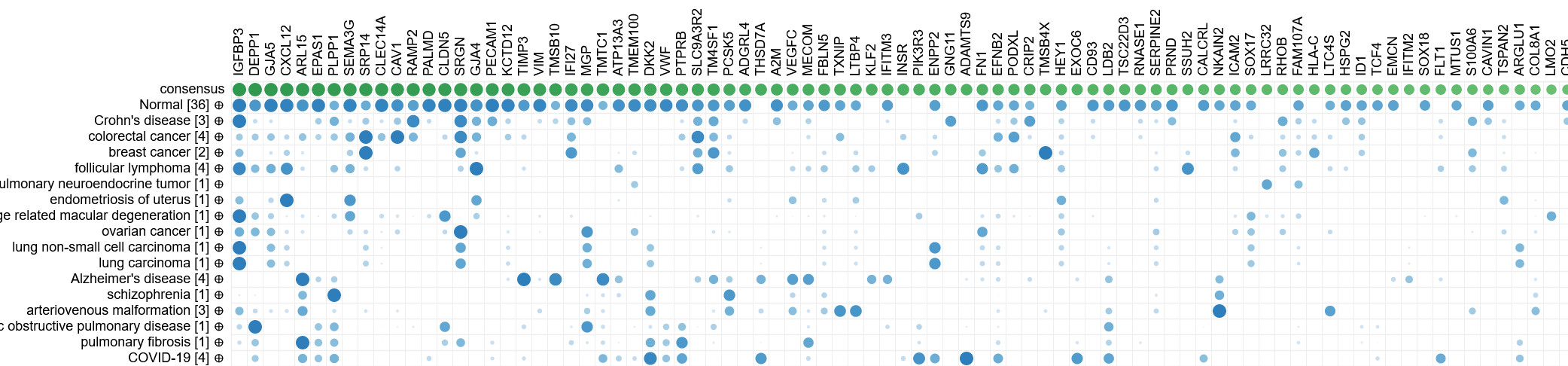
Biomarker discovery
Since CellKb calculates consensus signatures for all cell types across all tissues and conditions in which they are found,
it can be used to compare cell type markers between diseases or tissues to identify biomarkers that are common or unique to each condition.