Get, set, annotate - How AI cell type annotation methods compare to traditional approaches
This article explores commonly used methods for cell type annotation in scRNA-seq data. We examine traditional approaches such as CellTypist,
comparing them with AI methods like scGPT and SCimilarity. Additionally, we discuss how CellKb addresses the limitations of these
methods and enables rapid cell type annotation.
Read the article.
Released on: 21 April 2025
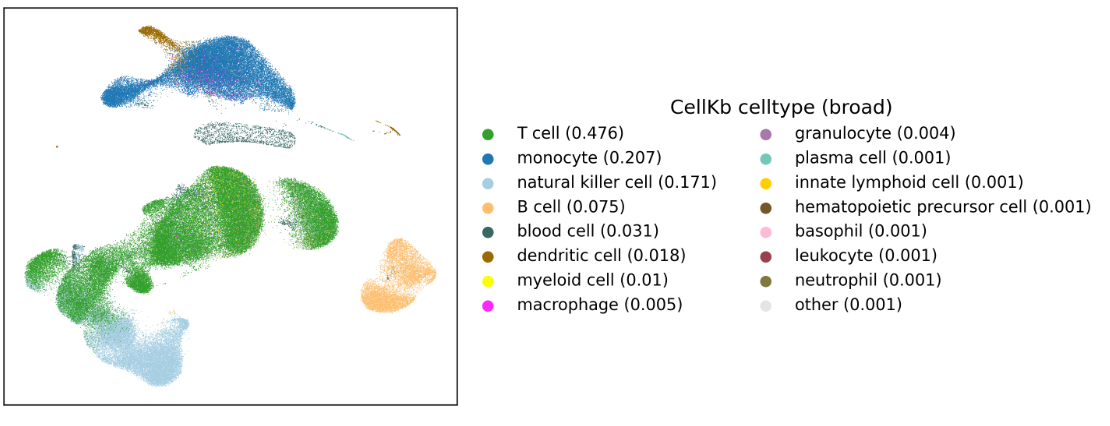
Annotating cells in the Tabula Muris Senis mouse liver dataset (CellKb vs CellTypist)
In this article, we assign cell types to cells in the Mouse liver dataset from the Tabula Muris Senis publication.
Each cell is assigned a cell type by CellKb individually, without clustering. We compare our results with the author annotations and annotations from CellTypist. Read the article.
Released on: 10 October 2024
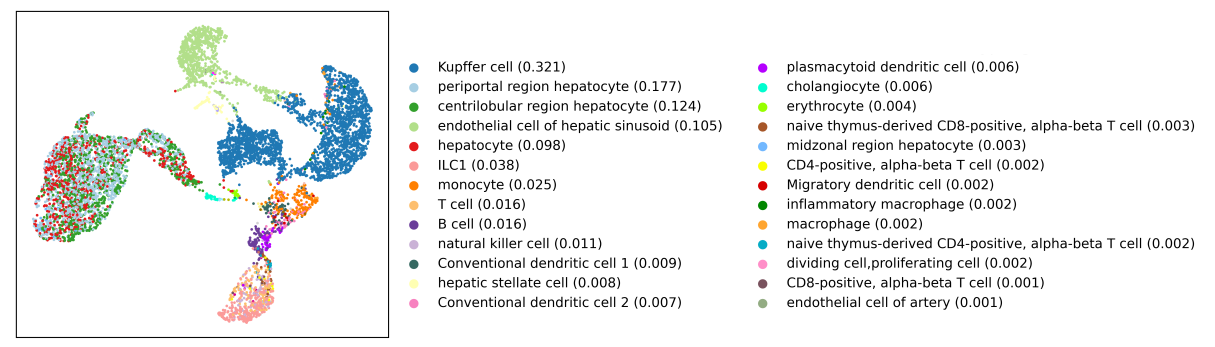
Annotating cells in the PBMC dataset (CellKb vs SingleR)
In this article, we assign cell types to cells in the Peripheral Blood Mononuclear Cells (PBMC) dataset. Each cell is assigned a cell type by CellKb individually, without clustering.
Annotating cells individually is more likely to identify rare cell types. Annotating cells individually will also give a better result when the clusters identified in your dataset are not cleanly separated. We compare our results with the results from SingleR. Read the article.
Released on: 11 January 2024
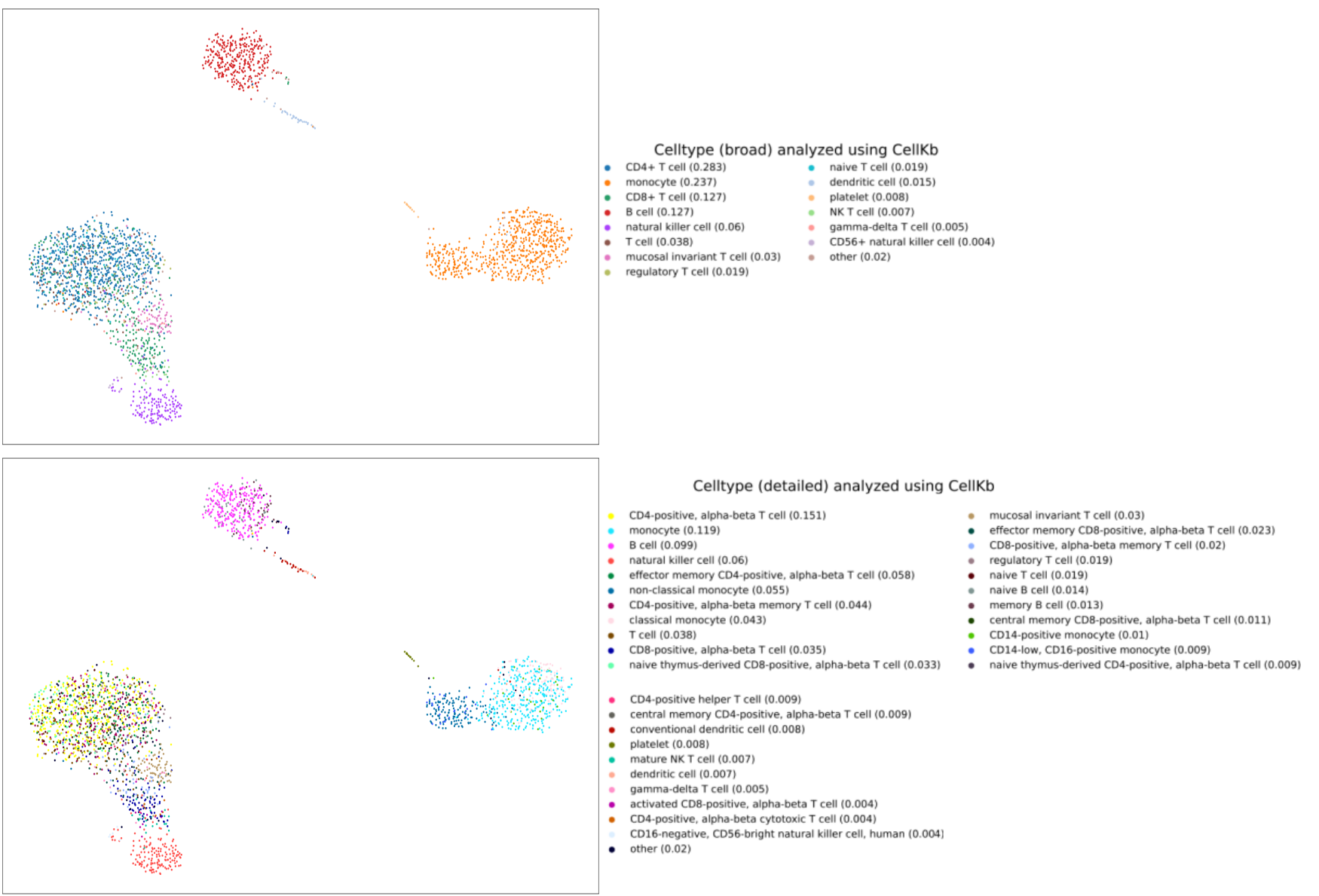
Cell type annotation of clusters in single-cell RNA-seq data
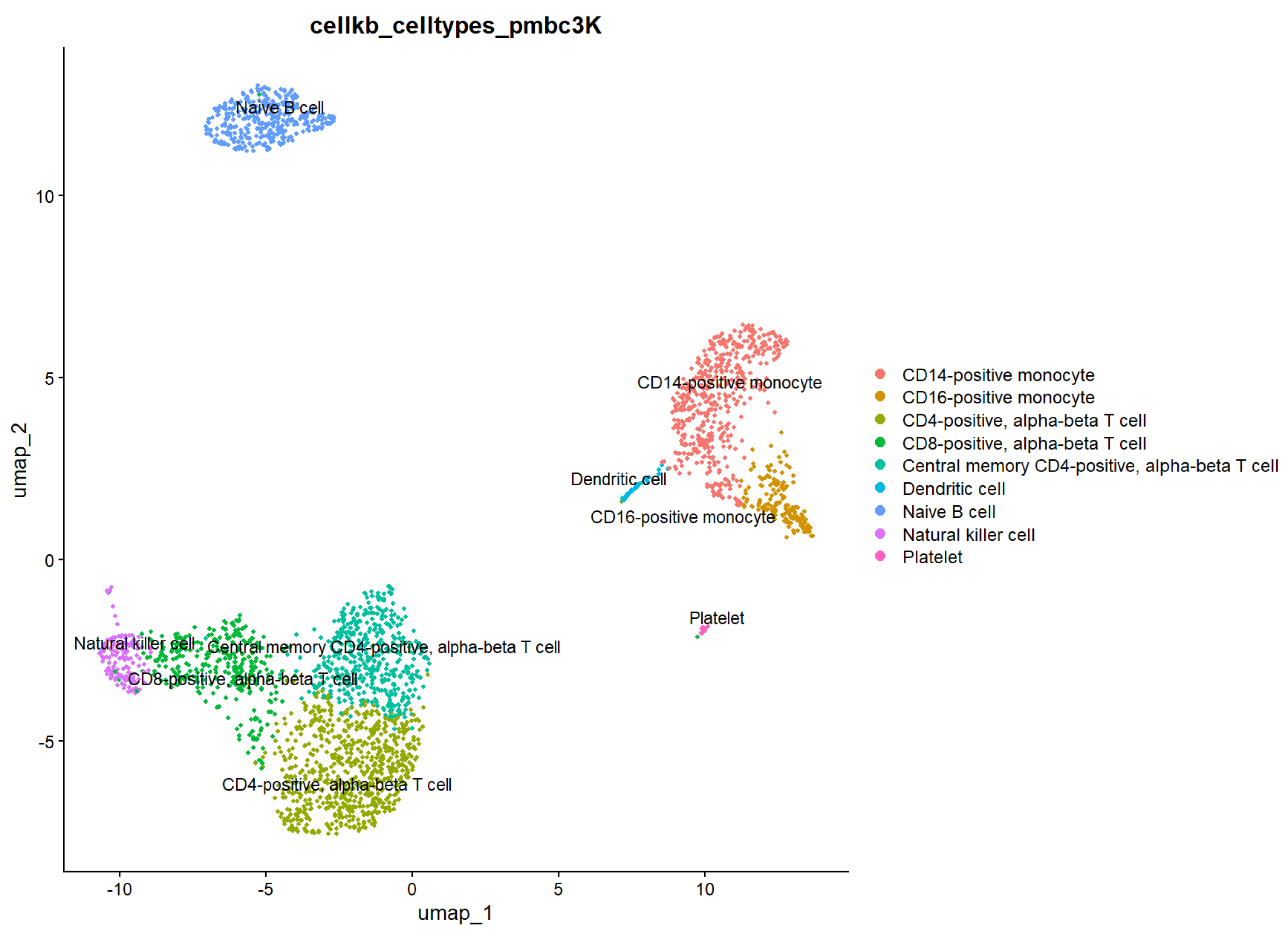
In this article, we assign cell types to clusters in the dataset of Peripheral Blood Mononuclear Cells (PBMC).
We identify the clusters and cluster-specific differentially expressed genes using Seurat. We then use these gene lists to identify the cell types using CellKb. We
compare our results with the results from other methods such as SingleR and PanglaoDB. Read the article.
Released on: 20 June 2023